According to unpublished data quoted by Inutsuka et al., SOB has been shown to be protective against DOX cardiac and renal toxicity in rats. Additionally, SOB exhibited synergy with DOX in colon-26 tumour-bearing mice as measured by prolonged survival of the animals. In the same study, the authors observed an increased G2/M population after incubating colon-26 cells with a combination of SOB and DOX for 24 h compared to DOX-treated samples. The G2/M arrest was also reported in a human mammary tumour cell line exposed to 4 mM SOB for 20 h. Given the gradual increase in ANT sensitivity as cells transition from relatively resistant G1 phase to G2/M phase, which was observed in exponentially growing Molt-4 cells, the accumulation of G2/M cells before ANT treatment could be beneficial as the relatively resistant population is depleted in favour of the more sensitive population. In our study, SOB alone did not induce significant G2/M phase arrest of HL-60 cells after treatment for 72 h. However, SOB significantly reduced the G1 and S phase populations, which could be favourable for increasing HL-60 cell sensitivity to DOX. The combination of SOB with DOX caused even more pronounced depletion of G1 and S phase cells together with an increase in polyploid cells and cell debris. Indeed, the combination of DOX or DAU with SOB in our study provoked unequivocally synergistic antiproliferative effects in HL-60 cells at all assessed concentrations and schedule. Conversely, SOB pre-incubation with NVCMs led to significant protection from DAU- and DOXinduced toxicity as assessed by LDH release, mitochondrial membrane potential measurements and caspase activity measurements. MER is a non-sedative barbiturate derivative that is structurally unrelated to bisdioxopiperazines. Its principal effect on replicating cells is the inhibition of chromosome condensation, activation of c-Jun and JNKs and induction of G2/M cell cycle blockade. In this study, although MER significantly protected NVCMs against both DAU- and DOX-induced LDH release, it also induced some toxicity in NVCMs. MER insignificantly reduced the DAU-induced activation of caspases and also only partially protected cardiomyocytes from DYm loss. However, of the three catalytic Vorinostat 149647-78-9 inhibitors examined in this study, MER showed the highest potential to act synergistically with ANTs in HL-60 cells, particularly at higher concentrations for which there was no tendency towards increased CI values. Additionally, with 3- or 6-h pre-incubations, the drug combinations remained consistently synergistic. The effects of MER on the cell cycle in HL-60 cells differed considerably from the bisdioxopiperazines. MER alone caused dramatic G2/M arrest, which was even more pronounced than that induced by DOX. In addition, the sub G1 and polynuclear populations increased. However, unlike DEX or SOB, the MER + DOX combination did not induce more pronounced cell cycle changes. As DEX and SOB are the members of the bis-dioxopiperazine family, they can be metabolized to 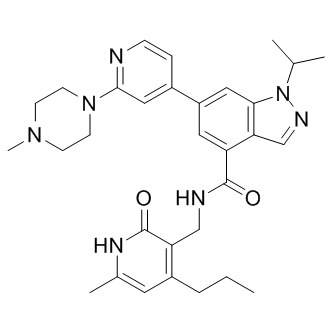 Fe-chelating compounds. Whereas DEX is well known to undergo metabolization to ADR925, SOB must be LY2109761 side effects probably first metabolized to ICRF-154 and then to the open-ring product, thus requiring two-step metabolism to metal chelator. To examine this issue, the Fe intracellular chelating properties of DEX, SOB and also MER were examined in H9c2 rat embryonic cardiomyoblast-derived cell line using the measurements of calcein-AM fluorescence intensity to compare the cardioprotection with their ability to chelate Fe. The experimental strong lipophilic Fe chelator SIH was repeatedly documented to chelate free Fe both in solution as well as in cells, and to displace Fe from its complex with calcein resulting in dequenching of its fluorescence and therefore it has been used as the reference chelator in our study. Indeed, fast increase in calcein fluorescence could be observed upon addition of 100 mM SIH to the cells with intracellular-trapped calcein-Fe complex and also DEX is able to significantly chelate Fe in comparison with control, although rather slowly and with only,30% efficiency as compared to SIH. The,2.5 h lag-time before the start of fluorescence increase was probably caused by the need of DEX hydrolysis to the chelating metabolite ADR-925. Similar result was observed by Hasinoff et al., who found that unlike ADR-925, which displaces Fe from the cell-trapped calcein-Fe complex in a pattern similar to SIH, DEX chelated Fe only partially and with substantial lag-time.
Fe-chelating compounds. Whereas DEX is well known to undergo metabolization to ADR925, SOB must be LY2109761 side effects probably first metabolized to ICRF-154 and then to the open-ring product, thus requiring two-step metabolism to metal chelator. To examine this issue, the Fe intracellular chelating properties of DEX, SOB and also MER were examined in H9c2 rat embryonic cardiomyoblast-derived cell line using the measurements of calcein-AM fluorescence intensity to compare the cardioprotection with their ability to chelate Fe. The experimental strong lipophilic Fe chelator SIH was repeatedly documented to chelate free Fe both in solution as well as in cells, and to displace Fe from its complex with calcein resulting in dequenching of its fluorescence and therefore it has been used as the reference chelator in our study. Indeed, fast increase in calcein fluorescence could be observed upon addition of 100 mM SIH to the cells with intracellular-trapped calcein-Fe complex and also DEX is able to significantly chelate Fe in comparison with control, although rather slowly and with only,30% efficiency as compared to SIH. The,2.5 h lag-time before the start of fluorescence increase was probably caused by the need of DEX hydrolysis to the chelating metabolite ADR-925. Similar result was observed by Hasinoff et al., who found that unlike ADR-925, which displaces Fe from the cell-trapped calcein-Fe complex in a pattern similar to SIH, DEX chelated Fe only partially and with substantial lag-time.
Monthly Archives: July 2019
The mutation Y253F and Y253H present on the P-loop is in close contact with imidazo pyridazine of ponatinib
The calculated free energies correlate with experimentally measured IC50 values and comparably ponatinib 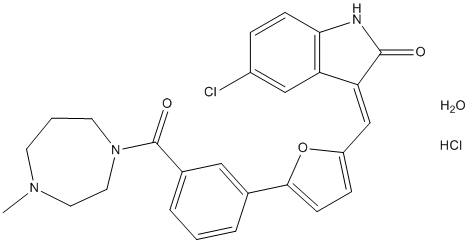 has better binding towards the mutation T315A than T315I. The free energy of BCR-ABLT315I Life Science Reagents complexed with imatinib is 29.89 kcal/mol indicating that ponatinib has higher binding towards T315I mutation compared to imatinib. Table 1 shows the distribution of electrostatic potential and contribution from neighbouring residues during MD simulations that are responsible for this free energy change. The Y253F mutation has 2 fold greater activity than Y253H, although the net SIE values for both complexes do not correlate with observed experimental values. These mutations show decrease in intermolecular coulomb energies compared to native kinase and Y253F mutation shows decreased vdW interaction energies. Phe317 located at middle of the hinge region is in ATP binding site, imidazo pyridazine ring of ponatinib interacts with Phe317 via pi-pi stacking and vdW contacts. From the analysis of MD simulations of F317V BCR-ABL kinase – ponatinib complex, we observed slightly increased intermolecular vdW energy and cavity value and decreased intermolecular coulomb value. The SIE free energy for F317V BCR-ABL kinase �C ponatinib complex is which is close to the SIE free energy of F317L. The residue Phe359 is located on turn region at the end of aChelix and is involved in the formation of a hydrophobic core with several residues from aC- helix including hydrophobic amino acids Val289 and Ile293. The F359V is adjacent to piperazine solubilization group of ponatinib and forms weak vdW interactions. The SIE binding free energy was observed for this complex. In spite of side chains being oriented away from the binding site of ponatinib, the P-loop mutations E255K and E255V are closer to ponatinib effecting its activity. The structure Enzalutamide superposition of ponatinib and imatinib is shown in Figure 4 and both inhibitors bind BCR-ABL kinase at exactly the same location. Further despite the peptide bond swap, both inhibitors vary only in two locations. 1. The presence of CF3 group on piperazine substituted phenyl ring. 2. The presence of acetylene linked imidazo pyridazine ring. The CF3 group makes close contacts with hydrophobic side chains of Leu298 and Leu354. From Table 2, we observed that Leu298 has stable interactions with CF3 throughout MD simulations in all mutations, while Leu354 experiences increased vdW interactions with mutations E255K and T315A. Thr315 is close to acetylene link of ponatinib and imidazo pyridazine ring is in a hydrophobic cavity that is enclosed by Leu248, Tyr253, Phe382, Phe317 and Leu370. Among these, Leu370 makes stable CH-pi interactions with imidazo pyridazine ring contributing to the stability of native and mutant complexes. The other four residues in most BCR-ABL kinase mutants when bound to ponatinib undergo high conformational changes during MD simulations. We believe that these conformational changes are responsible for ponatinib binding and inhibition of native and mutant BCR-ABL kinases. The pan-BCR-ABL kinase inhibitor, ponatinib is most popular for its inhibition of ABLT315I mutation at nano molar concentrations. Fourteen mutant ABL kinase structures complexed with ponatinib were modeled and we performed 25 ns of MD simulations to study the structural changes of protein when complexed with ponatinib within its binding site. Using the SIE method, we calculated binding free energies and its component of non bonding energies such as intermolecular vdW energies and reaction field energies. Further, coulomb and vdW contributions from individual amino acid residues in active site were calculated. The calculated SIE values are in the range 210.03 kcal/mol to 210.67 kcal/mol and correspond with the narrow range of IC50 values of native and mutant BCR-ABL kinase inhibition by ponatinib. From these MD simulations, we observed that fluctuations in residues from P-loop, b3-, b5- strands and aC- helix are mainly responsible for ponatinib binding to native and all mutant BCR-ABL kinases. Further, amino acid residues Met244, Lys245, Gln252, Gly254, Leu370 and Leu298 did not undergo any conformational changes due to mutations. The rest of the mutations effect ponatinib binding free energy calculations with its component energies evidently correlating with their activities.
has better binding towards the mutation T315A than T315I. The free energy of BCR-ABLT315I Life Science Reagents complexed with imatinib is 29.89 kcal/mol indicating that ponatinib has higher binding towards T315I mutation compared to imatinib. Table 1 shows the distribution of electrostatic potential and contribution from neighbouring residues during MD simulations that are responsible for this free energy change. The Y253F mutation has 2 fold greater activity than Y253H, although the net SIE values for both complexes do not correlate with observed experimental values. These mutations show decrease in intermolecular coulomb energies compared to native kinase and Y253F mutation shows decreased vdW interaction energies. Phe317 located at middle of the hinge region is in ATP binding site, imidazo pyridazine ring of ponatinib interacts with Phe317 via pi-pi stacking and vdW contacts. From the analysis of MD simulations of F317V BCR-ABL kinase – ponatinib complex, we observed slightly increased intermolecular vdW energy and cavity value and decreased intermolecular coulomb value. The SIE free energy for F317V BCR-ABL kinase �C ponatinib complex is which is close to the SIE free energy of F317L. The residue Phe359 is located on turn region at the end of aChelix and is involved in the formation of a hydrophobic core with several residues from aC- helix including hydrophobic amino acids Val289 and Ile293. The F359V is adjacent to piperazine solubilization group of ponatinib and forms weak vdW interactions. The SIE binding free energy was observed for this complex. In spite of side chains being oriented away from the binding site of ponatinib, the P-loop mutations E255K and E255V are closer to ponatinib effecting its activity. The structure Enzalutamide superposition of ponatinib and imatinib is shown in Figure 4 and both inhibitors bind BCR-ABL kinase at exactly the same location. Further despite the peptide bond swap, both inhibitors vary only in two locations. 1. The presence of CF3 group on piperazine substituted phenyl ring. 2. The presence of acetylene linked imidazo pyridazine ring. The CF3 group makes close contacts with hydrophobic side chains of Leu298 and Leu354. From Table 2, we observed that Leu298 has stable interactions with CF3 throughout MD simulations in all mutations, while Leu354 experiences increased vdW interactions with mutations E255K and T315A. Thr315 is close to acetylene link of ponatinib and imidazo pyridazine ring is in a hydrophobic cavity that is enclosed by Leu248, Tyr253, Phe382, Phe317 and Leu370. Among these, Leu370 makes stable CH-pi interactions with imidazo pyridazine ring contributing to the stability of native and mutant complexes. The other four residues in most BCR-ABL kinase mutants when bound to ponatinib undergo high conformational changes during MD simulations. We believe that these conformational changes are responsible for ponatinib binding and inhibition of native and mutant BCR-ABL kinases. The pan-BCR-ABL kinase inhibitor, ponatinib is most popular for its inhibition of ABLT315I mutation at nano molar concentrations. Fourteen mutant ABL kinase structures complexed with ponatinib were modeled and we performed 25 ns of MD simulations to study the structural changes of protein when complexed with ponatinib within its binding site. Using the SIE method, we calculated binding free energies and its component of non bonding energies such as intermolecular vdW energies and reaction field energies. Further, coulomb and vdW contributions from individual amino acid residues in active site were calculated. The calculated SIE values are in the range 210.03 kcal/mol to 210.67 kcal/mol and correspond with the narrow range of IC50 values of native and mutant BCR-ABL kinase inhibition by ponatinib. From these MD simulations, we observed that fluctuations in residues from P-loop, b3-, b5- strands and aC- helix are mainly responsible for ponatinib binding to native and all mutant BCR-ABL kinases. Further, amino acid residues Met244, Lys245, Gln252, Gly254, Leu370 and Leu298 did not undergo any conformational changes due to mutations. The rest of the mutations effect ponatinib binding free energy calculations with its component energies evidently correlating with their activities.
Ability to prevent aberrant FtsZ assembly at cell poles and ensures the integrity and dynamic nature of medial
Our findings suggest that the four domains of B.  subtilis EzrAhave separable, yet overlapping roles in mediating EzrA’s interaction with FtsZ and potentially other cellular RWJ 64809 factors. A summary of relevant phenotypes can be found in Figure 7. In particular, our data suggest the sole function of EzrA’s TM helix is to concentrate the protein at the plasma membrane where it can function most efficiently to inhibit aberrant FtsZ assembly at cell poles, promote the dynamic nature of the medial FtsZ ring, and to coordinate interactions between components of the cell division and cell wall synthesis machinery. Although deleting EzrA’s TM helix entirely led to an ezrA null phenotype, swapping EzrA��s TM helix with similarly oriented TM helices from either ZipA and FtsK, two E. coli cell division proteins, or TM helices from the E. coli respiratory proteins CccA and SdhA, had no discernable impact on EzrA function. All four EzrA TM chimeras exhibited wild type morphology with regard to cell size, cell division, FtsZ assembly and growth rate. These data raise questions about physiological relevance of BACTH data suggesting EzrA PI-103 interacts directly with a large number of almost exclusively extracellular proteins. One possibility is that such interactions are real, but dispensable for EzrA function in vivo. Alternatively, interaction between EzrA and primarily extracellular proteins may be artifacts of the BACTH assay itself. For example E. coli FtsZ may function as a bridge between EzrA and the target cell division proteins, bringing the T25 and T18 domains of adenylate cyclase into close enough proximity for synthesis of cyclic AMP. EzrA is known to inhibit assembly of E. coli FtsZ both in vivo and in vitro. Regardless of mechanism, the apparently generic nature of EzrA’s TM domain reinforces the need to obtain biochemical data confirming interactions identified between EzrA and components of the cell division machinery by BACTH. In contrast to the TM domain, our data suggests that EzrA’s four coiled-coils have specific and separable functions. CC1 and CC2 are required for EzrA mediated inhibition of FtsZ assembly at cell poles but dispensable for EzrA activity at midcell, while CC3 and CC4 appear to modulate interaction between EzrA and FtsZ throughout the cell. Deletion of CC1 and CC2or CC2 aloneresulted in a frequency of polar FtsZ rings approximately equivalent to that of an ezrA null mutant, but had little impact on EzrA localization to midcell or the stability of the medial FtsZ ring. We speculate that CC1 and CC2 play key roles in mediating interactions between EzrA and cytoplasmic components of the division machinery. For example the inability of the CC1 and CC2 deletions to inhibit polar FtsZ assembly may be due to loss of interactions between EzrA and proteins concentrated at the cell poles that normally help bring the division inhibitor into close proximity with FtsZ at this location. In contrast, ezrA mutants defective in CC3 or CC4 were resistant to overexpression of the minCD division inhibitor and also suppressed the heat sensitivity of the ftsZts allele, in addition to having a high frequency of polar FtsZ rings. Separable roles for CC1 and CC2 relative to CC3 and CC4 are further supported by our biochemical analysis of EzrA deletion mutants. In vitro, deletion of CC1 and CC2 in tandem or CC2 alone had little impact EzrA’s ability to interact with FtsZ, while deletion of CC3 and/or CC4 completely abolished interaction between EzrA and FtsZ. Based on these data we propose that CC1 and CC2 help coordinate interactions between EzrA and FtsZ specifically at the cell poles.
subtilis EzrAhave separable, yet overlapping roles in mediating EzrA’s interaction with FtsZ and potentially other cellular RWJ 64809 factors. A summary of relevant phenotypes can be found in Figure 7. In particular, our data suggest the sole function of EzrA’s TM helix is to concentrate the protein at the plasma membrane where it can function most efficiently to inhibit aberrant FtsZ assembly at cell poles, promote the dynamic nature of the medial FtsZ ring, and to coordinate interactions between components of the cell division and cell wall synthesis machinery. Although deleting EzrA’s TM helix entirely led to an ezrA null phenotype, swapping EzrA��s TM helix with similarly oriented TM helices from either ZipA and FtsK, two E. coli cell division proteins, or TM helices from the E. coli respiratory proteins CccA and SdhA, had no discernable impact on EzrA function. All four EzrA TM chimeras exhibited wild type morphology with regard to cell size, cell division, FtsZ assembly and growth rate. These data raise questions about physiological relevance of BACTH data suggesting EzrA PI-103 interacts directly with a large number of almost exclusively extracellular proteins. One possibility is that such interactions are real, but dispensable for EzrA function in vivo. Alternatively, interaction between EzrA and primarily extracellular proteins may be artifacts of the BACTH assay itself. For example E. coli FtsZ may function as a bridge between EzrA and the target cell division proteins, bringing the T25 and T18 domains of adenylate cyclase into close enough proximity for synthesis of cyclic AMP. EzrA is known to inhibit assembly of E. coli FtsZ both in vivo and in vitro. Regardless of mechanism, the apparently generic nature of EzrA’s TM domain reinforces the need to obtain biochemical data confirming interactions identified between EzrA and components of the cell division machinery by BACTH. In contrast to the TM domain, our data suggests that EzrA’s four coiled-coils have specific and separable functions. CC1 and CC2 are required for EzrA mediated inhibition of FtsZ assembly at cell poles but dispensable for EzrA activity at midcell, while CC3 and CC4 appear to modulate interaction between EzrA and FtsZ throughout the cell. Deletion of CC1 and CC2or CC2 aloneresulted in a frequency of polar FtsZ rings approximately equivalent to that of an ezrA null mutant, but had little impact on EzrA localization to midcell or the stability of the medial FtsZ ring. We speculate that CC1 and CC2 play key roles in mediating interactions between EzrA and cytoplasmic components of the division machinery. For example the inability of the CC1 and CC2 deletions to inhibit polar FtsZ assembly may be due to loss of interactions between EzrA and proteins concentrated at the cell poles that normally help bring the division inhibitor into close proximity with FtsZ at this location. In contrast, ezrA mutants defective in CC3 or CC4 were resistant to overexpression of the minCD division inhibitor and also suppressed the heat sensitivity of the ftsZts allele, in addition to having a high frequency of polar FtsZ rings. Separable roles for CC1 and CC2 relative to CC3 and CC4 are further supported by our biochemical analysis of EzrA deletion mutants. In vitro, deletion of CC1 and CC2 in tandem or CC2 alone had little impact EzrA’s ability to interact with FtsZ, while deletion of CC3 and/or CC4 completely abolished interaction between EzrA and FtsZ. Based on these data we propose that CC1 and CC2 help coordinate interactions between EzrA and FtsZ specifically at the cell poles.
PAI-1 is dispensable in pathological lymphangiogenesis in tumoral situations as well as in inflammatory disorders
The present study using genetic approaches provide for the first time evidences that in contrast to its pivotal role in pathological angiogenesis. This clearly demonstrates that distinct molecular pathways govern angiogenesis and lymphangiogenesis and that PAI-1 plays distinct roles in the remodelling of both circulation systems in pathological conditions. Providing that PAI-1 antagonists are used to inhibit angiogenesis, our results reveal that this approach will not have any effect on lymphangiogenesis. Although PAI-1 is dispensable for lymphangiogenesis, it is worth noting that other proteolytic systems are mandatory for this process and particularly matrix metalloproteases such as the MMP-2 whose deficiency impairs lymphangiogenesis in in vitro and in vivo models. Cyclin-dependent kinasesare poised to play a central role in the orderly transition of the eukaryotic cells through different stages of the mitotic cell division cycle. The activities of the CDKs are controlled by a tight network of regulatory mechanisms, which comprise activatory/inhibitory phosphorylation and dephosphorylation events, controlled degradation of the cyclin partner and association with effectors. Several CDKIsfunction as tumour supressorsand loss/subversion of its activitiesresults in the development of tumours, cancers and neoplasms. The importance of CDKIs in benign and malignant leukaemias, urological and other diseases is a subject of intense ongoing investigation. Though initially considered as tumour suppressors based on their ability to block cell proliferation, CDKIs play pertinent roles in the regulation of a myriad of cellular processes including transcription, apoptosis, cell migration and cytoskeletal dynamics, which may be oncogenic under certain circumstances. Due to the involvement of CDKs in critical cellular roles, inhibition of CDKs harbors immense relevance for anticancer therapy. Inhibition of CDKs could be accomplished both by over expression of cellular CDKIsas well as pharmacological inhibitors. Cellular CDKIs e.g. the tumour suppressor gene products p16INK4, p21WAF1, and p27KIP1, form the starting point for the design of mechanism-based CDK inhibitors. Analysis of the structural aspects of cellular CDKIs leads to the identification of inhibitory lead peptides amenable to peptidomimetic development. Conversion of these peptides into pharmaceutically useful molecules provides a wealth of potential drug candidates capable of inhibiting CDKs, blocking cell-cycle progression, modulating transcription and inducing apoptosis selectively in cancer cells. Some of these, such as flavopiridol, LDN-193189 7-hydroxystaurosporineand roscovitine, have  already reached the stage of ABT-199 clinical evaluation. These pharmacological CDKIs herald the opening of new avenues of clinical therapies against such intractable pathogens like human immunodeficiency virus and several protozoan parasites like Plasmodium, Trypanosoma and Leishmania. CDKIs also constitute potential targets for therapeutic stem cell manipulations. In the light of well established significance of CDKI proteins within the cell and in the development of pharmacological CDK inhibitors, it becomes essential to have facile methods of identifying these proteins. However, these exhibit a lot of diversity in their amino acid sequences. These are represented by INK4and Cip/Kipfamilies in mammals, Sic1 proteinin fungi and SIAMESE family and ICK/KRP family in plants. Owing to this enormous diversity, its identification is precluded by simple similarity-based approaches. As an alternative to similarity based methods, we applied two machine learning techniques.
already reached the stage of ABT-199 clinical evaluation. These pharmacological CDKIs herald the opening of new avenues of clinical therapies against such intractable pathogens like human immunodeficiency virus and several protozoan parasites like Plasmodium, Trypanosoma and Leishmania. CDKIs also constitute potential targets for therapeutic stem cell manipulations. In the light of well established significance of CDKI proteins within the cell and in the development of pharmacological CDK inhibitors, it becomes essential to have facile methods of identifying these proteins. However, these exhibit a lot of diversity in their amino acid sequences. These are represented by INK4and Cip/Kipfamilies in mammals, Sic1 proteinin fungi and SIAMESE family and ICK/KRP family in plants. Owing to this enormous diversity, its identification is precluded by simple similarity-based approaches. As an alternative to similarity based methods, we applied two machine learning techniques.
The treatment of influenza virus-infected patients is mainly based on increasing resistance against currently approved
We used compositional features including amino acid composition, Split Amino Acid Composition, Di-peptide Compositionand gapped dipeptide composition, as well as evolutionary information from the Position-Specific Scoring Matrixprofiles obtained from Position-Specific Iterative-Basic Local Alignment Search Toolfor training the classifiers. Given the immense biomedical merit and therapeutic potential of CDKIs, we anticipate that this would be a useful tool for the research community. Three iterations of PSI-BLAST were carried out at an E-value threshold of 0.001. Each sequence was used as the query sequence once while the rest were used as the reference database and this was looped over each sequence. It was found that 10 sequences did not find any significant hit, bringing forth that general methods of similarity-based searches do not provide a reliable solution to the identification of CDKIs and a method specific to these proteins should be developed. Therefore, we set forth to explore machinelearning based methods based on various protein features for the prediction of CDKI proteins. PSSM-based SVM classifiers are have been employed for a plethora of classification problems in biology and are well known for their remarkable performance for extremely diverse proteins like lipocalins, nucleic acid binding proteins, etc. Apart from capturing residue composition, the PSSM profiles encapsulate useful information about conservation of residues at crucial positions within the protein sequence, because in evolution the amino acid residues with similar physico-chemical properties tend to be highly conserved due to selective pressure. This is the first report of a machine-learning-based method for identification of CDKI protein sequences. Previously, such approaches have been applied to the computational identification of other components of the cell cycle including cyclinsand CDK phosphorylation substrates. Our tool simply represents a complementary tool to allow the detection of CDK inhibitors, since we prove that PFAM signatures miss a significant number of the already known CDK inhibitors from the Silmitasertib PKC inhibitor non-redundant set. However such machine learning based methods do come at the cost of some false positive predictions, which should be as minimal as possible. We tested this on an independent dataset comprising of randomly picked up nonCDKIs as well as kinases and phosphatases which are the most likely candidates for false positive predictions and indeed obtained a low false positive prediction rate. In this study, we observed that SVM based methods are more efficient than ANN in discrimination of CDKI and non-CDKI sequences, despite the imbalance in the size  of the positive and the negative training datasets. This was observed with all the types of features. For an experimenter, a judicious approach would be minimizing the number of CDKIs to be characterized by increasing the threshold to higher SVM score, in order to get only the topmost candidates for further work. Supplementing these with other complementary evidence like domain WZ8040 EGFR/HER2 inhibitor knowledge and sub-cellular localization may provide inroads to the discovery of novel CDKIs and further our understanding of cell cycle regulation and other cellular phenomena. In future, the availability of more sequences and inclusion of more features may further enhance the prediction accuracy. While the current H1N1 influenzapandemic was ongoing in 2010, efforts were made to develop new antiviral agents for influenza treatment that possess an improved spectrum of activity or better pharmacologic profiles, compared to current treatments.
of the positive and the negative training datasets. This was observed with all the types of features. For an experimenter, a judicious approach would be minimizing the number of CDKIs to be characterized by increasing the threshold to higher SVM score, in order to get only the topmost candidates for further work. Supplementing these with other complementary evidence like domain WZ8040 EGFR/HER2 inhibitor knowledge and sub-cellular localization may provide inroads to the discovery of novel CDKIs and further our understanding of cell cycle regulation and other cellular phenomena. In future, the availability of more sequences and inclusion of more features may further enhance the prediction accuracy. While the current H1N1 influenzapandemic was ongoing in 2010, efforts were made to develop new antiviral agents for influenza treatment that possess an improved spectrum of activity or better pharmacologic profiles, compared to current treatments.