To screen proteins related to the progression of gastric carcinogenesis, we also compared non-neoplastic gastric tissues with GC samples of individuals with and without lymph node metastases. For the selection of differentially expressed proteins, we used a Mechlorethamine hydrochloride statistical parametrics test with bootstrapping resampling. We have undertaken a comprehensive computational analysis of tissue proteomic data to discover pathways and networks involved in gastric oncogenesis and progression. The possible associations among enolase 1 and heat shock 27 kDa protein 1 gene and protein expression, and clinicopathological characteristics were also evaluated in individuals from Northern Brazil. These two proteins have been reported frequently as deregulated molecules in previous GC proteomic studies in other populations. However, they were never evaluated in a Brazilian population. Although some proteomic-based studies were previously performed in human primary gastric tumors, to our knowledge, only three studies focus on noncardia GC. Most GC proteomic studies identified differentially expressed proteins based only on a fold change between two conditions. Other previous GC proteomic studies performed statistical analyses to compare the protein expression between groups, but without controlling the type I  error. It is interesting to note that we compared the protein profiling of tumors and non-neoplastic samples using parametric tests with bootstrapping for differentially expressed protein identification. The resampling methods, commonly used in microarray studies, have been used to make p-value adjustments for multiple testing procedures which control the FWER and take into account the dependence structure between test statistics. The objective is to create many sets of bootstrap samples by resampling with the replacements from the original data. This supports the current data and indicates that the findings should be further investigated to identify or validate clinically relevant targets for the diagnosis and/ or the prognosis of GC. The identified proteins were grouped into different classes based on functional information available. Most of the identified proteins were intracellular organelle proteins and were part of the membrane-bound organelles, especially the mitochondria. These proteins are involved mainly in cellular metabolic processes and oxidation reduction. The comparison of tumors with lymph node metastases and control samples revealed the transport and the establishment of location as Butenafine hydrochloride enriched biological processes. This was not observed in the comparison of tumors without lymph node metastases and control samples. The oxidoreductase activity followed by coenzyme binding activity were the main molecular functions of the proteins involved in gastric carcinogenesis. Concerning the chromosomal location, most of the proteins were encoded by genes located at chromosome 1, chromosome 19, and chromosome 11. Complex karyotypes of gastric tumors preferentially involve these chromosomes, as well as chromosomes 3, 6, 7, 8, 13 and 17. Gain and loss of chromosome regions of 1, 19 and 11 were previously reported by our research group in GC samples. Thus, these chromosomes may contain several loci with dosage-sensitive genes. We have undertaken a comprehensive computational analysis of tissue proteomic data to discover pathways and networks involved in gastric oncogenesis and progression. Table 2 shows the principal canonical pathways using the Ingenuity Pathway Analysis database. The principal canonical pathways in the gastric carcinogenesis process were involved in the energy metabolism pathways including mitochondrial dysfunction, pyruvate metabolism, oxidative phosphorylation, citrate circle, and glycolysis/gluconeogenesis.
error. It is interesting to note that we compared the protein profiling of tumors and non-neoplastic samples using parametric tests with bootstrapping for differentially expressed protein identification. The resampling methods, commonly used in microarray studies, have been used to make p-value adjustments for multiple testing procedures which control the FWER and take into account the dependence structure between test statistics. The objective is to create many sets of bootstrap samples by resampling with the replacements from the original data. This supports the current data and indicates that the findings should be further investigated to identify or validate clinically relevant targets for the diagnosis and/ or the prognosis of GC. The identified proteins were grouped into different classes based on functional information available. Most of the identified proteins were intracellular organelle proteins and were part of the membrane-bound organelles, especially the mitochondria. These proteins are involved mainly in cellular metabolic processes and oxidation reduction. The comparison of tumors with lymph node metastases and control samples revealed the transport and the establishment of location as Butenafine hydrochloride enriched biological processes. This was not observed in the comparison of tumors without lymph node metastases and control samples. The oxidoreductase activity followed by coenzyme binding activity were the main molecular functions of the proteins involved in gastric carcinogenesis. Concerning the chromosomal location, most of the proteins were encoded by genes located at chromosome 1, chromosome 19, and chromosome 11. Complex karyotypes of gastric tumors preferentially involve these chromosomes, as well as chromosomes 3, 6, 7, 8, 13 and 17. Gain and loss of chromosome regions of 1, 19 and 11 were previously reported by our research group in GC samples. Thus, these chromosomes may contain several loci with dosage-sensitive genes. We have undertaken a comprehensive computational analysis of tissue proteomic data to discover pathways and networks involved in gastric oncogenesis and progression. Table 2 shows the principal canonical pathways using the Ingenuity Pathway Analysis database. The principal canonical pathways in the gastric carcinogenesis process were involved in the energy metabolism pathways including mitochondrial dysfunction, pyruvate metabolism, oxidative phosphorylation, citrate circle, and glycolysis/gluconeogenesis.
Monthly Archives: June 2019
Towards a loop distribution in which genes become distal to the NM as a function of time also holds for neurons
However, the term RS actually implies three separate phenomena that are not causally related: there is a RS that correlates with the progressive shortening of chromosome telomeres as a function of number of cell divisions, but this phenomenon has only be observed in cells from primates. A second kind of RS known as STASIS may occur prematurely as a response to diverse cellular stressors, this STASIS depends on specific genetic functions and so it can be reverted by mutation or inactivation of such genetic functions. There is a third kind of RS that occurs stochastically and so it increases its probability as a function of time, this seems to occur in all cells both in vitro and in vivo and it is not reversible. A self-stabilizing model for DNA-NM interactions as a function of age, in which a stochastic but time-dependent process leads to a significant increase of DNA-NM interactions, resulting in an integral and highly-stable structural system, has been proposed for explaining a common physical basis for both stochastic RS senescence and the post-mitotic state. We have recently shown that aged hepatocytes and early post-mitotic neurons have similar highly-stable NHOS. The present work 3,4,5-Trimethoxyphenylacetic acid expands such result by showing that continued stabilization of the NHOS occurs in post-mitotic neurons even after the fourth post-natal week when according to microanatomical criteria they formally become TD. The fact that the continued stabilization of the neuronal NHOS as a function of time has no overt impact on gene expression in neurons, suggests that this phenomenon do not depends on functional constraints. The DNA of each chromosome constitutes a continuous double-stranded fibre in which each strand has a rigid helical backbone resulting from the strong phosphodiester bonds between the deoxyribose sugars of thousands of nucleotides along the strand, whereas the weak hydrogen bonds between the nitrogenous bases in the anti-parallel strands can be broken and reestablished quite easily. Thus the torsional stress of the long DNA molecule along its axis might be dissipated by breaking the hydrogen bonds between both strands, yet by looping and supercoiling along its axis DNA can dissipate the stress without compromising its structural integrity. Hence the interactions DNA-NM that result in a large number of structural DNA loops in the interphase nucleus may be a natural answer to a structural stress problem posed by the intrinsic configuration of DNA. This phenomenon is independent of proteins that constitute chromatin and apparently depends on DNA-NM interactions by means of so-called indirect Atropine sulfate readouts. Nevertheless, the local chromatin conformation may have a role in determining the choice of DNA sequences available for interaction with the NM in vivo, since chromatin proteins may compete or hinder such DNANM interaction. A DNA loop pattern in which most genes lie close to the actual MARs attached to the NM is observed in baby and young neurons, similarly to what was reported in hepatocytes of equivalent age, but this is a highly  improbable distribution considering that protein-coding genes are rare within the genome and so most potential MARs are not close to any protein-coding gene. Yet in the hepatocytes this anomalous distribution changes in time to one in which most genes are distal to the NM, this correlates with a trend towards shortening and homogenization of the average DNA- loop size as a function of age. In aged neurons most target genes studied also become distal to the NM indicating.
improbable distribution considering that protein-coding genes are rare within the genome and so most potential MARs are not close to any protein-coding gene. Yet in the hepatocytes this anomalous distribution changes in time to one in which most genes are distal to the NM, this correlates with a trend towards shortening and homogenization of the average DNA- loop size as a function of age. In aged neurons most target genes studied also become distal to the NM indicating.
Slowing of this Gln transport resulting in longer residence times in the GluGln Cycle would also lead to higher effective
This suggests that further exploration of regional Glu and Glx in the brain in ASD is Cinoxacin warranted. One brain region frequently implicated in ASD, but where glutamatergic metabolites have been little explored, is the anterior 3,4,5-Trimethoxyphenylacetic acid cingulate cortex. Evidence from several investigative modalities points to involvement of the anterior cingulate in ASD, including neuropathology, structural MRI, fMRI, MRS, PET, SPECT, and EEG evoked potentials. One hypothesis also relates anterior cingulate dysfunction to deficits in joint attention and social orienting in ASD. This plentiful prior work gives reason to search for further abnormalities, perhaps involving the glutamatergic system, in anterior cingulate. Here, we report on two independent studies of glutamatergic neurometabolites in the anterior cingulate cortex in pediatric ASD; the first a pilot study, the second a larger follow-up investigation. Within the cingulate gyrus, our investigations focused on the pregenual anterior cingulate cortex subregion, one of the eight subregions in Vogt’s definitive parcellation of the human cingulate cortex. Most of the above-cited neuroimaging studies localized their acquisition or analysis volumes based on the older four-subregion or two-subregion cingulate models. As the eight-subregion model is most consistent with extant neuropathological, neuroimaging, and  neurocognitive data, we anticipated that focusing on a subregion within this model would improve odds of detecting Glx effects and would permit more anatomically standardized statement of our results. Moreover, recent neuroimaging investigations, including multimodal MRS-fMRI and combined fMRI and genetic work, of autistic symptoms and autistic traits in healthy subjects have demonstrated focal effects within the pACC, raising the possibilities for finding MRS effects there as well. Consequences of chronic excess Glx could include abnormal development, ongoing excitotoxic cell damage, and inefficient utilization of cell energy. The small pilot investigation, Experiment 1, indicated elevated Glx in pACC in ASD. A larger follow-up study, Experiment 2, in which the sample with ASD and the control sample were now better matched for gender and IQ again found elevated Glx in ASD, albeit restricted to right pACC. Thus, two separate studies using independent samples of children with ASD and controls and scanning on two different systems both support the idea of pACC hyperglutamatergia in ASD. As discussed in our review, prior MRS studies reporting Glu or Glx in ASD have yielded mixed results, presumably due to differences in subject samples and MRS methods. Using rigorous MRSI methods, Friedman et al. found no effects of ASD on Glx in the cingulate. Unfortunately, these authors do not specify which subregion of the cingulate they sampled. Also, all the subjects with ASD in their study underwent propofol sedation at time of scan, whereas that was the case for only one subject in Experiment 1 and for none in Experiment 2. Shulman et al. point out that brain energetic metabolism, and thence the rate of Glu-Gln cycling and thereby possibly Glx levels, is intimately related to anesthesiainduced level of consciousness. Finally, the subjects in Friedman et al. were considerably younger than ours. Anatomic neuroimaging suggests differences between ASD in early and later childhood; for example, the well-known observation of brain overgrowth in ASD between 2�C 4 years that arrests or normalizes in later childhood and adolescence.
neurocognitive data, we anticipated that focusing on a subregion within this model would improve odds of detecting Glx effects and would permit more anatomically standardized statement of our results. Moreover, recent neuroimaging investigations, including multimodal MRS-fMRI and combined fMRI and genetic work, of autistic symptoms and autistic traits in healthy subjects have demonstrated focal effects within the pACC, raising the possibilities for finding MRS effects there as well. Consequences of chronic excess Glx could include abnormal development, ongoing excitotoxic cell damage, and inefficient utilization of cell energy. The small pilot investigation, Experiment 1, indicated elevated Glx in pACC in ASD. A larger follow-up study, Experiment 2, in which the sample with ASD and the control sample were now better matched for gender and IQ again found elevated Glx in ASD, albeit restricted to right pACC. Thus, two separate studies using independent samples of children with ASD and controls and scanning on two different systems both support the idea of pACC hyperglutamatergia in ASD. As discussed in our review, prior MRS studies reporting Glu or Glx in ASD have yielded mixed results, presumably due to differences in subject samples and MRS methods. Using rigorous MRSI methods, Friedman et al. found no effects of ASD on Glx in the cingulate. Unfortunately, these authors do not specify which subregion of the cingulate they sampled. Also, all the subjects with ASD in their study underwent propofol sedation at time of scan, whereas that was the case for only one subject in Experiment 1 and for none in Experiment 2. Shulman et al. point out that brain energetic metabolism, and thence the rate of Glu-Gln cycling and thereby possibly Glx levels, is intimately related to anesthesiainduced level of consciousness. Finally, the subjects in Friedman et al. were considerably younger than ours. Anatomic neuroimaging suggests differences between ASD in early and later childhood; for example, the well-known observation of brain overgrowth in ASD between 2�C 4 years that arrests or normalizes in later childhood and adolescence.
Although this description prevents from separate tuning of similarity measures in the protein and ligand spaces
Therefore, we concluded that this predicted drug-target interaction network tends to be controlled by only a small number of drugs and targets, which have a lot of available pharmacological interaction information in the learning dataset. Finding new therapeutic indications for the existing drugs represents an efficient parallel approach to the drug discovery, since existing drugs already have extensive clinical history and toxicological information. All above models and the derived information show that new potential drug-target interaction can be effectively predicted by our proposed approach. And to achieve this goal, i.e., to further predict the novel targets for the existing drugs by using our models, two representative small molecules MDMA and Resveratrol were selected presently to illustrate the models’ applications, since the comprehensive drug-target interaction network is immensely huge. MDMA is a known psychoactive drug, which is also effective in the treatment of post-traumatic stress disorder. And Resveratrol has the potential of creating anti-inflammatory and anticancer effects. The selection of these two molecules is owing to that their related target information has been reported in literature, but has not been included in training set of the obtained RF Model I and RF Model II. The potential targets of these two molecules are predicted from the pool of all 3987 target proteins using the RF Model I and RF Model II, respectively. Traditional drug discovery is largely based upon ‘one moleculeone target-one disease’ model, but there is a growing recognition that drugs work by targeting multiple proteins. The biological network and pathways possessing inherent redundancy and robustness imply that regulating a single target might fall short of producing the desired therapeutic effects. Therefore, the development of multiple drug-target interaction prediction models to investigate disease-associated drug-target network will undoubtedly be an enduring trend for future drug discovery. In this report, by integrating the information from the chemical structure, protein sequence and pharmacological drug-target interaction data, we developed a set of in silico models using a large-scale dataset to predict the potential drug-target interactions. All models were evaluated and verified by both internal and external validations. The outcomes demonstrated the strength of our proposed method for predicting drug-target interaction, which indicates that the conserved binding patterns between drugs and targets can be extracted by our approach from the dataset that contains adequate feature vectors for chemical-protein pairs. Selecting a suitable encoding of the Dexrazoxane hydrochloride compounds and proteins information is one of the main computational challenges for the prediction of drug-target interactions using in silico tools. In our case, we apply  DRAGON molecular descriptors and structural and physicochemical properties descriptors to represent ligands and targets, respectively. Our successful predictions indicate that this adopted chemical and proteins encoding can effectively distinguish the drug-target binding pairs from the non-binding pairs. Additionally, the choice of merging protein and ligand descriptors into a single Ginsenoside-Ro vector describing both partners was also adopted in this study, which means that the structural similarity between the two different drugs/targets are independently evaluated by the same measure and are then multiplied to give the overall similarity.
DRAGON molecular descriptors and structural and physicochemical properties descriptors to represent ligands and targets, respectively. Our successful predictions indicate that this adopted chemical and proteins encoding can effectively distinguish the drug-target binding pairs from the non-binding pairs. Additionally, the choice of merging protein and ligand descriptors into a single Ginsenoside-Ro vector describing both partners was also adopted in this study, which means that the structural similarity between the two different drugs/targets are independently evaluated by the same measure and are then multiplied to give the overall similarity.
Under such condition for identifying the true neuronal nuclei in both N1 and N2 populations
The DNA loops plus the NM constitute a nucleoid, a very large nucleoprotein aggregate generated by lysis of nuclei at pH 8.0 in non-ionic detergent and the presence of high salt concentration. In nucleoids the DNA remains essentially intact but it lacks the nucleosome structure because of the dissociation of histones and other chromatin proteins; yet the naked DNA loops remain topologically constrained and supercoiled being attached to the NM. Nucleoids are a standard preparation for evaluating the NHOS. After treating the N1 and N2 populations for extraction of nucleoids the resulting preparations are highly enriched in NeuN-positive neuronal nucleoids while the presence of NeuN-negative nucleoids from glial cells is significantly reduced, indicating that the glial-cell nuclei are poorly resistant to the NMextraction conditions. This fact underlines the overall higher stability of the NHOS from neurons when compared to that of glial cells. Moreover, we have recently shown that NeuN/ Fox-3 is an intrinsic component of the neuronal NM and so it is a reliable marker of the neuronal NHOS. Nucleoids are also known as nuclear halos since exposure of such structures to DNA-intercalating agents like ethidium bromide leads to unwinding of the DNA loops that form a fluorescent DNA halo around the NM periphery. The EB acts as a molecular lever causing the unwinding of loop DNA and this process induces strong tearing forces that impinge upon the NM as the DNA rotates and expands during unwinding. Thus the stability of the DNA-NM interactions and of the NM itself can be evaluated by titrating the response of the nucleoids to increasing concentrations of EB since weak DNA-NM interactions are eliminated at lower EB concentrations. Hence, exposing the corresponding nucleoids to a high concentration of EB leads to maximum halo expansion and under such conditions any significant change in the distribution of the DNA-NM interactions or any significant weakness in the strength of such interactions is revealed as distortions and heterogeneities in the resulting halo Gomisin-D surrounding the NM. Moreover, a weak NM can be fractured by the tearing forces resulting from DNA loop unwinding an expansion. After exposure to high the nucleoids from neurons of the four post-natal ages studied display well defined DNA halos that surround the undisturbed NM framework. This is in contrast with nucleoids from P0 and P7 hepatocytes that are destroyed by the resulting EB-induced DNA unwinding while only the nucleoids from P80 and P540 hepatocytes are able to withstand the effects of EB-induced DNA unwinding, 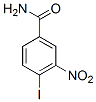 in a similar fashion as the neuronal nucleoids. We determined the average diameter of both nucleus and NM in neurons from all post-natal ages studied. The results indicate that the NM diameter is greater than the corresponding nuclear diameter at all ages studied, suggesting that the neuronal NM is resilient as it has been observed in lower eukaryotes. The average DNA halo size has been correlated with the average DNA-loop size, therefore by measuring the average DNA-halo radius after maximum halo expansion it is possible to estimate the average DNA-loop size. The results indicate that there is no real significant change in the average DNA loop size in neurons from maturity to old age. This is in contrast to what we have observed in hepatocytes in which the average DNA loop size is reduced by 36% from P80 to P540. Lomitapide Mesylate Nevertheless, the average DNA loop size in neurons from all ages studied is significantly shorter than that in aged P540 hepatocytes.
in a similar fashion as the neuronal nucleoids. We determined the average diameter of both nucleus and NM in neurons from all post-natal ages studied. The results indicate that the NM diameter is greater than the corresponding nuclear diameter at all ages studied, suggesting that the neuronal NM is resilient as it has been observed in lower eukaryotes. The average DNA halo size has been correlated with the average DNA-loop size, therefore by measuring the average DNA-halo radius after maximum halo expansion it is possible to estimate the average DNA-loop size. The results indicate that there is no real significant change in the average DNA loop size in neurons from maturity to old age. This is in contrast to what we have observed in hepatocytes in which the average DNA loop size is reduced by 36% from P80 to P540. Lomitapide Mesylate Nevertheless, the average DNA loop size in neurons from all ages studied is significantly shorter than that in aged P540 hepatocytes.