We identified a structurally diverse set of small scaffolds that may be overlaid onto distinct regions of the c64 ligand as present in the crystal structure, and our M. tuberculosis H37Rv active sets were searched for hits that contain these substructures. Twelve such 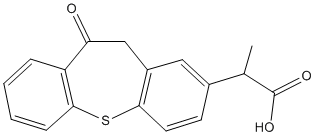 small scaffolds led to the ASP1517 clinical trial identification of forty-three compounds among the reported TB actives, and these are shown in Figure S1. Out of the M. tuberculosis H37Rv actives identified based on these searches thirty-three compounds were selected for evaluation against Mt-GuaB2. Compound 7759844 was docked into the Mt-GuaB2-IMP complex structure using the InducedFit docking protocol from the Schrodinger software package that treated residues within 5 A ? of docked ligand conformers as flexible. Two possible docked orientations were predicted for this ligand, one showed excellent consistency with the limited SAR data available for this lead scaffold, as listed in Table 3. Therefore, we considered this mode as our working model of 7759844 binding to Mt-GuaB2, for further validation. The potential of IMPDH for immunosuppressive, cancer and antiviral chemotherapy has been previously explored. Recently, Mt-GuaB2 has gained attention as a promising anti-mycobacterial target, associated with a number of distinct inhibitor scaffolds. The applicability of available bacterial IMPDH inhibitors may be limited by toxicity issues as there is an important human homolog, and lead optimization must focus on selectivity as well as potency. In the search for new lead compounds as potential MtGuaB2 inhibitors, we employed a BU 4061T Proteasome inhibitor designed scaffold-based approach utilizing the IMPDH crystal structure with c64 cocrystallized in the active site. M. tuberculosis H37Rv active sets from our previous screens were searched based on a set of designed scaffolds listed in Figure S1 as well as a known IMPDH scaffold. Of the identified M. tuberculosis actives, we evaluated thirty-three compounds for their potential to inhibit Mt-GuaB2 functional activity and for their inhibitory potency against M. smegmatis. In the present study, the assay was adapted to a 200 ml volume, conditions were optimized by varying concentrations of the substrate IMP, the cofactor NAD+and the enzyme, and the Km were determined for IMP. The optimized assay yielded a reaction that proceeded linearly over a 5 min period. Next, we studied the inhibition of Mt-GuaB2 by a series of commercially available compounds, primarily from the Chembridge and Life Chemicals libraries. When compared to the DPU’s, these compounds show an improvement in enzyme inhibitory activity, yielding low micromolar inhibitors. The Ki values were obtained for all inhibitors with respect to both substrates IMP and the patterns of inhibition were inferred from the graphs. Among the Chembridge compounds, four exhibited low micromolar affinity Ki values. Out of the Life Chemicals compound set, F1374-1083 showed a low micromolar inhibition constant. All inhibitors from the Chembridge compound set showed an uncompetitive pattern of inhibition against both substrates. Uncompetitive inhibition has been observed previously versus both IMP and NAD +, in the case of compounds having a strong preference for the E-XMP* complex. The observation of an uncompetitive pattern of inhibition also depended on assay conditions. In the case of uncompetitive inhibition, both the substrates bind to the enzyme before the inhibitor binds. The uncompetitive inhibitors have an advantage as drugs since inhibition increased due to accumulation of substrates. The post-translational modification of core histones plays a central role in epigenetic gene regulation. Modifications are put in place by families of modifying and demodifying enzymes.
small scaffolds led to the ASP1517 clinical trial identification of forty-three compounds among the reported TB actives, and these are shown in Figure S1. Out of the M. tuberculosis H37Rv actives identified based on these searches thirty-three compounds were selected for evaluation against Mt-GuaB2. Compound 7759844 was docked into the Mt-GuaB2-IMP complex structure using the InducedFit docking protocol from the Schrodinger software package that treated residues within 5 A ? of docked ligand conformers as flexible. Two possible docked orientations were predicted for this ligand, one showed excellent consistency with the limited SAR data available for this lead scaffold, as listed in Table 3. Therefore, we considered this mode as our working model of 7759844 binding to Mt-GuaB2, for further validation. The potential of IMPDH for immunosuppressive, cancer and antiviral chemotherapy has been previously explored. Recently, Mt-GuaB2 has gained attention as a promising anti-mycobacterial target, associated with a number of distinct inhibitor scaffolds. The applicability of available bacterial IMPDH inhibitors may be limited by toxicity issues as there is an important human homolog, and lead optimization must focus on selectivity as well as potency. In the search for new lead compounds as potential MtGuaB2 inhibitors, we employed a BU 4061T Proteasome inhibitor designed scaffold-based approach utilizing the IMPDH crystal structure with c64 cocrystallized in the active site. M. tuberculosis H37Rv active sets from our previous screens were searched based on a set of designed scaffolds listed in Figure S1 as well as a known IMPDH scaffold. Of the identified M. tuberculosis actives, we evaluated thirty-three compounds for their potential to inhibit Mt-GuaB2 functional activity and for their inhibitory potency against M. smegmatis. In the present study, the assay was adapted to a 200 ml volume, conditions were optimized by varying concentrations of the substrate IMP, the cofactor NAD+and the enzyme, and the Km were determined for IMP. The optimized assay yielded a reaction that proceeded linearly over a 5 min period. Next, we studied the inhibition of Mt-GuaB2 by a series of commercially available compounds, primarily from the Chembridge and Life Chemicals libraries. When compared to the DPU’s, these compounds show an improvement in enzyme inhibitory activity, yielding low micromolar inhibitors. The Ki values were obtained for all inhibitors with respect to both substrates IMP and the patterns of inhibition were inferred from the graphs. Among the Chembridge compounds, four exhibited low micromolar affinity Ki values. Out of the Life Chemicals compound set, F1374-1083 showed a low micromolar inhibition constant. All inhibitors from the Chembridge compound set showed an uncompetitive pattern of inhibition against both substrates. Uncompetitive inhibition has been observed previously versus both IMP and NAD +, in the case of compounds having a strong preference for the E-XMP* complex. The observation of an uncompetitive pattern of inhibition also depended on assay conditions. In the case of uncompetitive inhibition, both the substrates bind to the enzyme before the inhibitor binds. The uncompetitive inhibitors have an advantage as drugs since inhibition increased due to accumulation of substrates. The post-translational modification of core histones plays a central role in epigenetic gene regulation. Modifications are put in place by families of modifying and demodifying enzymes.
The activities of which are influenced by local concentrations of metabolites or the small molecule c46 cocrystallized
Leave a reply